Brain tumour is one of the most fatal types of cancer that affects humans. It’s the biggest cancer killer of children and adults under 40, amounting to 14 million new cancer cases per year. And unfortunately causes 8,8 million deaths globally. These statistics are horrifying, but the good news is that deep learning is helping on a large scale to enhance brain tumour treatment.
This is the subject Lars Sjösund, former Senior AI Research Engineer at Peltarion and currently, AI Research Engineer at NAVER Copr, presented on at the Nordic Data Science and Machine Learning Summit 2017. He talked about a joint project they did where they used deep learning for brain tumour segmentation without a big dataset.


Deep learning and brain tumour segmentation
The radiation is one of the therapy choices most commonly used because it’s least invasive. In the radiation treatment, high radiation sources such as gamma rays or X-rays are used to target the tumour tissue, killing the cancer cells and saving healthy ones.
In order to be able to perform radiation treatment, doctors have to first create a detailed map of the patient’s brain which is called brain tumour segmentation. However, the process very complex and time-consuming. As Lars puts numbers to context, for a head and neck tumour, doctors need 4-6 hours to perform brain segmentation, while for a brain tumour, it takes around 1 hour.
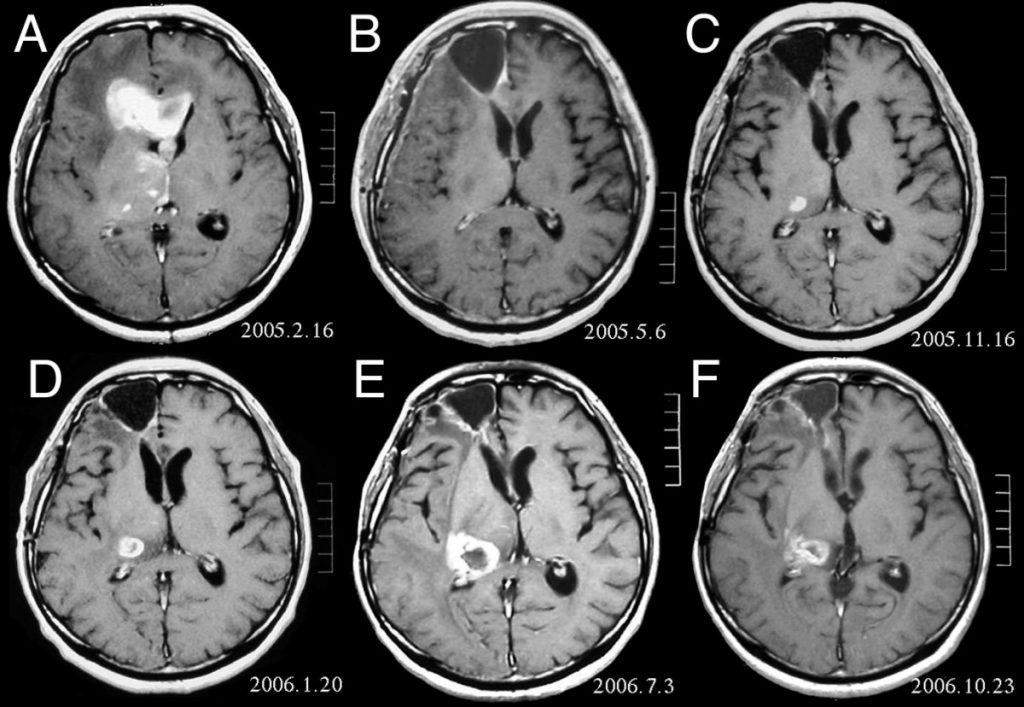

“If we want to scale up the radiation treatment to 50% of the population, we would need 10,000 doctors working full-time on only segmentation”, states Lars. And this is where deep learning can help by reducing the time doctors spend on segmentation.
Deep learning can contribute to enhancing the quality of the segmentation. As it depends on the skills of the doctor, the average similarity score between two doctors is 85% which is worrying. “We can help take the skills from the more experienced doctors and transfer them to less experienced ones,” says Lars.
If we want to scale up the radiation treatment to 50% of the population, we would need 10,000 doctors working full-time on only segmentation.
The dataset
The dataset Peltarion used for the project was an open-source that comes with a multi-model brain tumour segmentation challenge. It consisted of 276 patients with a brain tumour, and for all patients, they had 4 different 3D MRI scans which served as input data. They also had a target segmentation map with 4 different tumour classes. Their goal was to create a model that takes the patients’ MRI scans and outputs the target segmentation map.
And here they encountered the first challenge. “We only had 276 samples. We asked ourselves if it would enough to create a useful model,” says Lars.


We only had 276 samples. We asked ourselves if it would enough to create a useful model.
Models used in brain tumour segmentation
Peltarion used deep neural networks, which spared them from doing feature engineering and they could feed the almost raw data into the network so it learns good representation.
For this purpose, they made use of classical architecture for unsupervised learning called Autoencoder. It works by feeding the input in the encoder part, and after some kind of encoder representation, it is fed into the decoder part, which then outputs a new image. They trained the architecture the minimise the difference between the input and the output image. “But then the question arises why would we like to create this complicated copying machine,” recounts Lars.
The middle part between the encoder and the decoder part is a bottleneck layer which contains much fewer neurons than the number of pixels in the input image, he explains. This forces the encoder to compress the image and create a high-level of representation. And then the decoder learns how to decompress this high level of representation and produce the output image.
Peltarion had an idea to transform this model into a model that could do segmentation by switching the output target from trying to recreate the input to creating the segmentation map. But it only sounded feasible in theory, while in practice it didn’t work at all.
They turned to the U-net model, which has the Autoencoder structure as the base, but there are also skip connections to the output. So the input can flow both through the Autoencoder but it can also skip some layers. By using this model, the decoder can not only use the high-level representation to create segmentation, but also use low-level representations.
The U-net architecture today has a vast use in segmentation and the biological image field.


The results of the model training
The initial results Peltarion got were really satisfying, with 97.6% pixel-level accuracy. But when they looked at the qualitative results, they figured out that their networks didn’t learn anything. After doing the training, the networks predicted that there are no tumours anywhere. But why were their results so diagonally different?
When they looked into the problem, they saw that they have a huge class imbalance with 97.6% of non-tumour pixels and only 2.4% of tumour pixels. So by analogy, their network could be almost 100% correct by predicting that there was no tumour at all, which is catastrophically wrong.
To solve the problem, they changed what the network focused on. In their first training effort, being wrong on a tumour pixel was equally bad as being wrong on a non-tumour pixel. The second time they indicated that being wrong on a tumour pixel was much worse than being wrong on a non-tumour pixel. And after training the network again they got much better results. The predictions looked much more similar to the target.
When comparing how well doctors and the U-net neural network did, Lars points out that neural network had almost the same similarity score as the doctors’’. But we should emphasize again that Peltarion got these results by training on only 276 data points. How was it possible to get such good results with so few data points?

One possible solution Lars indicates is that how much data you need is domain-dependant. For example, when we compare two data sets – a tumour data set and an image data set, we come to the realisation that the tumour data sets are structurally very similar, so to cover a large part of all possible variations, we don’t need that much data. While for the image set, all data points are very different and to cover a large part of variations of natural images, we would need a lot more data points.
Taking into consideration Peltarion’s results, Lars advises companies that are asking themselves how many data points they need to build a model, should instead ask themselves if they have enough data to cover a large part of the possible variations of the domain they are trying to solve the problem in.
Challenges
When discovering new frontiers, challenges are inevitable. Some of the challenges Lars mentioned they encountered in their project were:
- Reproducibility – Reproducing the model and results may prove difficult because they relied on many open-source tools. Additionally, the neural networks they were using were that much developed.
- Fast-moving dependencies – The open-source tools they used tend to move and change pretty quickly, and getting consistent results is hard.
- Bookkeeping of results and configurations – Keeping track of what configuration led to what results, how different parameter settings affected results, version control of models, data sets, code&configurations and results is a challenge with these type of projects. Also, the tools for bookkeeping are still immature, Lars emphasises.
Companies that are asking themselves how many data points they need to build a model, should instead ask themselves if they have enough data to cover a large part of the possible variations of the domain they are trying to solve the problem in.
Future outlook
But the biggest challenge that this kind of deep learning projects face is the scarcity of data and small data sets, which is often the case in the medical field, Lars points out. The main reason is privacy concerns. But another reason is that labelled data in the medical field is very expensive. But there is a large body of unlabelled data which can be used in combination with labelled data which considerably improves learning and results. The goal of Peltarion with future projects is to combine unlabelled data with labelled data and explore semi-supervised learning.














Add comment